Introduction
Soil is a dynamic living matrix as well as a crucial resource for agricultural products. Aggregates of organic matter is accumulated in the rhizospheric zone. Because of microbial activities and high population in the zone, it has attracted much interest [1]. Gram-positive PGPR taxa that include Corynebacterium, B. cereus and B. thuringiensis. Gram- negative PGPR are fluorescent and non-fluorescent pseudomonads (P gladioli and P cepacia) and various members of the Enterobacteriaceae. The genus Bacillus represents one of the most abundant and phylogenetically diverse groups of easily cultivable PGPR [2]. Bacilli, due to their avid rhizosphere colonization and PGP characteristics, offer considerable interest for improving crop productivity and yield [3,4]. Along with 16S rRNA gene sequencing, is often employed for identification and molecular typing of bacterial species. It involves amplification of the BOX-elements (interspersed repetitive DNA sequences present in bacterial genomes) with BOX-A1R primer and demonstrates intraspecies diversity [5,6]. Bioinformatics is a science of retrieving and utilizing biological data for identification, characterization and typing of all kinds of pathogens [7,8]. Its application has been so enormous and revolutionary especially for clinical diagnosis, treatment, detection of virulence, resistance as well as outbreak investigation. In this study, using our knowledge in molecular biology and bioinformatics combined with basic knowledge in microbiology. We isolated PGPR from a farmland by using the conventional method of media cultivations, extracted the DNA, run polymerase chain reaction and sequenced to obtain nucleotides sequences.
Materials and methods
Procedures for the isolation and selection of isolates from the rhizosphere
One gram of [1g] of every rhizospheric soil sample was homogenized into 9.0 ml of sterile distilled H2O supplemented with 0.025. this was vortexed for 10 minutes during a serial dilution of soil sample. This translated to 1.0 m each of 103 and105 dilution which was inoculated on sterile agar plate using pour plate method. The plates were swirled and permitted to solidify. The plates were incubated for 18 hours and biochemical test carried out.
Biochemical Assay: Tentative Identification of Bacterial
Motility test
Clean glass slides were taken and placed on the table with depletion top, a canopy slip was taken and wax was applied on its four corners. A loopful of culture was transferred exactly at the middle of the duvet slips. Cavity slide was placed on the duvet slip and pressed gently. The preparation was lifted gently in order that the culture is suspended and was observed under 40 X objective of microscope.
Gram stain
Thin smear of bacterial culture was made on clean glass slide, it was air dried and warmth fixed, the Smear was covered with Gentian violet for 30 seconds, The slide was washed with water Smear was covered with Grams Iodine solution for 60 seconds, The Slide was washed with 95% ethyl alcohol and washed again with water, the smear was covered with safranin for 30 seconds, Washed with water and blot dried, air dried and observed under microscope.
Catalase test
Nutrient agar medium was prepared and the medium was poured into culture tubes. It was sterilized by autoclaving at 151b pressure for quarter-hour. The agar slants were inoculated with test organisms an inoculated agar slant was kept as control the cultures were incubated at 35°C and 3-4 drops of Peroxide was added on the expansion of every slant culture. The culture was observed for the presence or absence of gas bubbles.
Oxidase test
A piece of paper was divided into three equal sections and labeled with the name of organism. A loop-full of the culture was rubbed on the moistened paper employing a sterile loop. The colour of the smear was checked exactly between 15-30 seconds after rubbing the cells on the reagent moistened paper. A deep blue colour indicated positive reaction while light violet or purple colour developed within 10 seconds was recorded as negative.
Indole production test:
1% tryptone broth was prepared and sterilized using autoclave at 151 bs for fifteen minutes. The tryptone broth was inoculated with test organism and an uninoculated tube was kept as control. The tubes were incubated at 35°C for 48 hours. About 1ml of Kovac’s reagent was added for 48 hours of incubation. The tubes were shaken gently after intervals of 10 to 15 minutes; the tubes were allowed to stay for jiffy to allow the reagent to return to the highest while the tubes were observed for cerise layers.
Citrate utilization test
Simon’s citrate agar media was prepared and sterilized by autoclaving under standard condition. 5ml of the media was poured into the culture tubes and slants were prepared. In Simon’s citrate agar, slants were inoculated with test organisms. The uninoculated tubes were kept as control, the tubes were incubated at 370C for 48 hours.
Starch hydrolysis test
Starch agar media was prepared and sterilized using autoclave at 151 bs for 15 minutes the media was poured into petri dish plate and allowed to solidify the test organism was inoculated on to the plate with a sterile transfer loop the plate was incubated at 35°C for 48 hours after incubation the plate was flooded with Gram’s Iodine plate was observed for clear zone round the test organism.
Nitrate production test
Trypticase Nitrate broth was prepared broth was sterilized alongside glassware using autoclave at 151 bs for 15 minutes the broth was cooled and poured into the tube the test organism was inoculated using sterile loop the tubes were incubated at 37°C for 24-48hours after incubation 2-3 drops of reagent A-sulfanilic acid and equal amount of reagent B-Dimethyl a-naphthalamine was added to the broth culture the broth was observed for the looks of red colour absence of red colour showed negative effect of nitrate reduction it had been confirmed by adding pinch of zinc dust and therefore the tube was shaken vigorously appearance of red color confirmed the zinc had reduced the nitrate and therefore the organism was negative for nitrate reduction.
DNA Extraction Procedure
Bacterial DNA preparation it designed by JENA bioscience for easy and fast isolation of genomic DNA from both gram-positive and gram negative bacteria sample. The solution based system reduce DNA fragmentation that may be problematic in spin column or filtration based technique because phenol or chloroform is not use. It is safe and does not produce any harmful waste. Solution based genomic DNA purification it guarantee minimal DNA fragmentation and yield DNA size up to 150kb.
Preparation procedure
For S pack (50 preps): Before start, add 500 μl dd-water to the Proteinase K tube, 125 μl dd-water to the Lysozyme tube, 150 μl. Add-water to the RNase A tube and 48 ml 96-99 % Ethanol (not included in the kit) to the Washing Buffer bottle. Buffer has a resuspension of PP-214S 50 prep of 16 mls was put into PP-214L 250 of 80 ml. Lysis Buffer of 16 ml was dissolved in 80ml. Also, Binding Buffer 16 m80 ml, RNase A (50 mg/ml) was diluted in 7.5mg pp in 5x 7.5 mg and Lysozyme(100 mg/ml) of 12.5 mg was diluted in 5x 12.5 mg. Proteinase K(10 mg/ml of 5 mg of 5x 5 mg was added into activation Buffer 6 ml into 30 ml of pp and washing buffer. 48 ml of Ethanol (final volume 60ml) was added into 120 ml of Ethanol to each bottle (final volume 150 ml each) and Elution Buffer of 5 m into l25 ml. It is essential to use the correct amount of starting material in order to obtain optimal DNA yield. A maximum amount of 108 bacteria cells can generally be processed at a time. Overnight cultured bacteria cells can also be used. Cell pellets can be stored at -70 °C for several months until used [9].
Identification of PCR PGPR Using 16SrRNA Gene Sequencing
DNA template was prepared by picking a private colony and dissolving it in 1X Tris- EDTA solution [10]. Partial 16S rRNA gene sequences of the strains was acquired after PCR amplification of the genes by using the protocol described by Katsivila [11], using universal forward and reverse primers: 9F (5′- GAGTTTGATCCTGGCTCAG-3′) and 1510R (5′-GGCTACCTTGTTACGA-3′). Reaction mixture (50 μL) was denatured at 94⁰C for two min, followed by primer annealing at 55⁰C for 1 min and primer extension at 72⁰C for two min and eventually extension was obtained at 72⁰C for 10 min during a thermocycler [12-14]. Separation of amplified PCR products of 16S ribosomal gene was administered on 1% agarose gel in 0.5X TE (Tris- EDTA) buffer containing 2 μl ethidium bromide marker. The gel was examined under UV light and photographed by gel documentation system. The purified PCR products were sent to MACROGEN (Seoul, Korea) for sequencing by using four universal forward and reverse primers. The PCR products was analyzed of the selected isolates based on Amplified Ribosomal DNA Restriction Analysis (ARDRA) profiles by automated florescent dye terminator method (Sanger sequencing method). The results of sequence attained from MACROGEN, Korea were BLAST in program of the EzTaxon server [15] and therefore the sequences of closely related species were retrieved to determine the precise nomenclature of the bacterial isolates. The sequences were aligned with the relevant sequences retrieved from GenBank by using CLUSTAL W 1.8 XP program within the MEGA 5.2 software [16,17]. Aligned sequences were analyzed using an equivalent of the same software to construct unrooted phylogenetic trees by using the neighbor-joining method with bootstrap values supported 1000 replications [18].
Sequence Accession Numbers
The 16S rRNA gene sequences obtained in this study were deposited in the GenBank under accession numbers MW362290.1-MW362295.1 [22].
Results
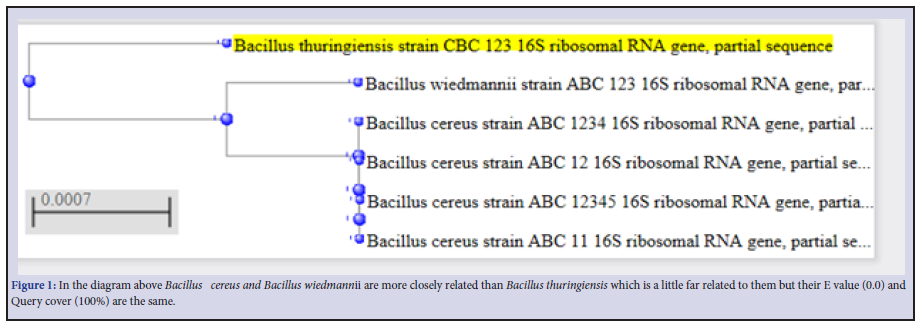
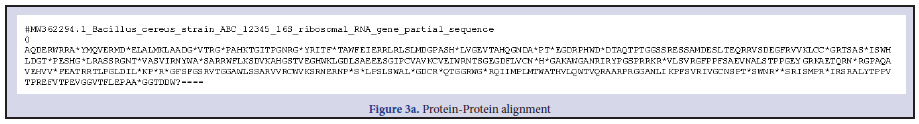
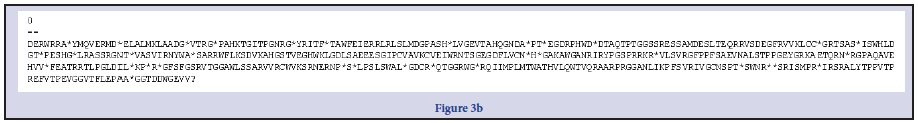
Three Bacillus strains were isolated from the rhizospheric regions of the Rice, pepper, sugar cane and vegetables. All the isolates were strong fermenters on EMB, weak on MacConkey, strongly positive on MRS, SSA and moderately positive on Centrimid agar. The cells; when viewed under microscope and under immersion oil, showed strong motility, non-indole Gram-positive rod shaped Table 1. To further strengthen identification after the preliminary identification based on morphological characteristics, molecular identification for amplification of essential genes; the isolates were subjected to PCR and subsequent sequencing techniques. The resultants nucleotides sequences were submitted in GenBank for accession numbers. This was further blasted on NCBI using BLASTn for homology and evolution relatedness using Mega X software (Figure 1). B. thuringensis (MW 362295.1) was used as the query against the other strains of Bacillus during BLAST for homology detection. Both nucleotides sequences and protein sequences were used for the alignment as obtained in Figure 2 a-f and Figure 3 a-e respectively. A phylogenetic tree, constructed based on the 16S rRNA gene sequences, indicated a considerable genetic homogeneity among the six Bacillus isolates (Figure1).
Discussion
The plant growth promoting abilities of bacteria isolates from the rhizosphere of four vegetative were examined. These three isolates were identified as Bacillus wiedmannii, Bacillus cereus and bacillus thuringiensis using 16SrRNA.
Colonial and morphological Characteristics
The colonial characteristics of all the bacterial isolate in this study is in conformity with early works by [19-21] was achieved from the observation of the macroscopic colonies of each of the isolate on different and differential media.
Biochemical characteristics of Bacterial Isolate
Biochemical test are among the most important method for microbial identification [19]. Routine biochemical test that can be used to ensure accuracy of identification of an unknown bacteria isolate include test for indole test, catalase test lipase test Gram stain.
16S rRNA gene-based molecular characterization
Amplicons of approximately 1500 bp were obtained after PCR amplification of the 16S rRNA. NCBI-BLAST analysis of the 16S rRNA gene sequences of all the test Rhizobacteria isolates (GenBank accession numbers. MW362291, MW362292, MW362293, MW362294, MW362295, MW362296 indicated that all six isolates are Bacillus spp., sharing 99–100% similarity with members of the genus Bacillus [22,23].
Phylogenetic Tree
Phylogenetic tree in this study showed evolution relatedness of the Bacillus spp. using the Neighbor-Joining method [18,22] with B. thuringensis was used for out grouping.